Advancing nanopore squiggle interpretation through SquiDBase data centralisation and algorithmic benchmarking
This poster was presented at the Revolutionizing Next-Generation Sequencing conference (6th edition, March 2025), organized by VIB Tools and Technologies.
Abstract
Nanopore sequencing is a groundbreaking technology for DNA and RNA analysis that directly reads nucleic acids, generating ionic current signals (squiggles) that capture both nucleotide sequences and epigenetic modifications. Its advantages include portability to resource-limited areas, the ability to detect long DNA fragments, and the capacity to preserve epigenetic information without requiring additional wet-lab techniques. Despite these benefits, the conventional sequencing workflow relies on GPU-intensive basecalling, which demands substantial computational resources and discards epigenetic data by focusing solely on canonical bases. Most researchers store basecalled FASTQ files, losing the rich information contained in squiggles.
To harness the untapped potential of squiggle data, we launched https://squidbase.org/, which is a centralized data repository designed to store raw nanopore signals from microorganisms and viruses, addressing the lack of curated repositories for squiggle data. Unlike traditional repositories such as the Sequence Read Archive, SquiDBase provides access to both legacy R9 and recent R10 datasets, promoting FAIR data and facilitating algorithm development.
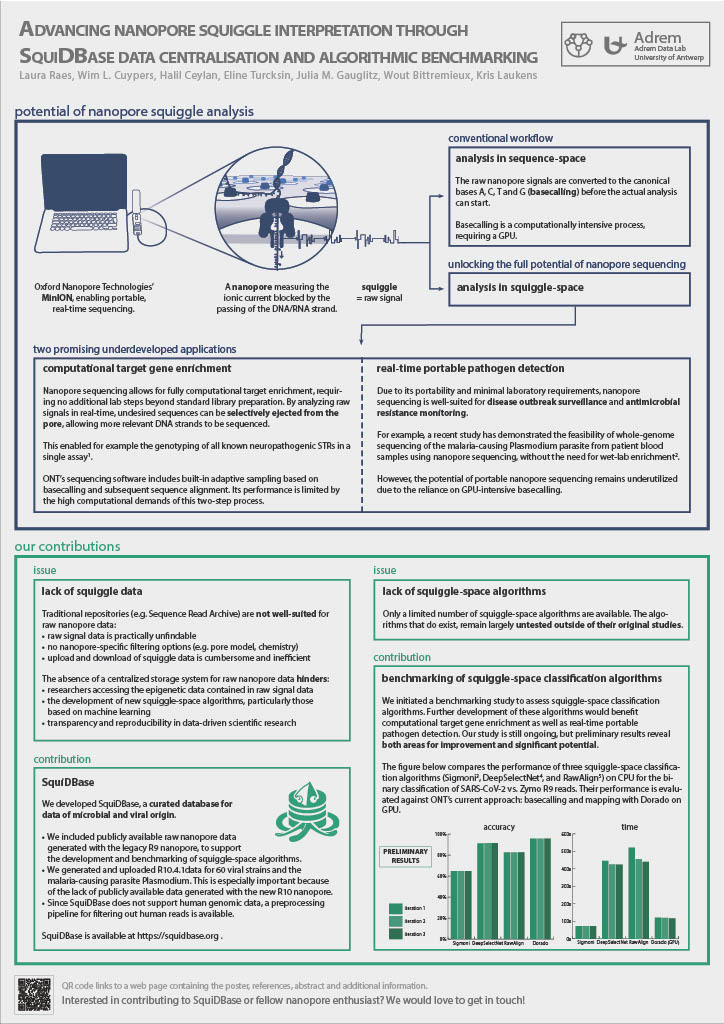
Additionally, we initiated a benchmarking study to assess squiggle-space classification algorithms that analyze raw ionic current data. These algorithms enable real-time enrichment of target regions (adaptive sampling) and rapid identification of sample composition, crucial for pathogen monitoring. However, their performance outside original studies remains largely untested. Our initial findings indicate that existing algorithms underperform on recent R10 data, demonstrating the added value of SquiDBase in supporting further advancements.
Additional information
SquiDBase
SquiDBase can be found at SquiDBase.org. If you have any questions about SquiDBase or would like to contribute viral or microbial data, feel free to contact us!
Remarks concerning our preliminary benchmarking results
- The tools were tested on a dataset comprising of:
- 20,000 reads with a length of at least 4500 signals, randomly selected from the CADDE Centre Sars-CoV2 R9 dataset.
- 20,000 reads with a length of at least 4500 signals, randomly selected from the Loman Lab Zymo-GridION-EVEN-BB-SN dataset.
- The minimal read length requirement of 4500 signals originates from the DeepSelectNet tool: this tool discards reads shorter than this threshold.
- The training data for DeepSelectNet consisted of 20,000 + 20,000 reads (> 4500 signals) from the same datasets.
- There was no overlap between the training and testing data.
- All tools were benchmarked on CPU.
- The performance of the tools was compared to Oxford Nanopore Technologies’ current approach: basecalling + mapping.
- For this we used their tool Dorado, and ran it on GPU.
- To determine Dorado’s accuracy, we threw away reads that were split by Dorado, non-primary alignments and supplementary alignments.
References
- Igor Stevanovski et al., Comprehensive genetic diagnosis of tandem repeat expansion disorders with programmable targeted nanopore sequencing. Sci. Adv. 8, eabm5386 (2022). DOI:10.1126/sciadv.abm5386
- De Meulenaere K, Cuypers WL, Gauglitz JM, Guetens P, Rosanas-Urgell A, Laukens K, Cuypers B. 2024. Selective whole-genome sequencing of Plasmodium parasites directly from blood samples by nanopore adaptive sampling. mBio15:e01967-23. doi.org/10.1128/mbio.01967-23
- Vikram S Shivakumar, Omar Y Ahmed, Sam Kovaka, Mohsen Zakeri, Ben Langmead, Sigmoni: classification of nanopore signal with a compressed pangenome index, Bioinformatics, Volume 40, Issue Supplement_1, July 2024, Pages i287–i296, doi.org/10.1093/bioinformatics/btae213
- Senanayake, A., Gamaarachchi, H., Herath, D. et al. DeepSelectNet: deep neural network based selective sequencing for oxford nanopore sequencing. BMC Bioinformatics 24, 31 (2023). doi.org/10.1186/s12859-023-05151-0
- Lindegger J, Firtina C, Ghiasi NM, Sadrosadati M, Alser M, Mutlu O: RawAlign: Accurate, Fast, and Scalable Raw Nanopore Signal Mapping via Combining Seeding and Alignment. doi.org/10.48550/arXiv.2310.05037